Global Sensitivity Analysis of the Lotka-Volterra model
The tutorial covers a workflow of using GlobalSensitivity.jl on the Lotka-Volterra differential equation. We showcase how to use multiple GSA methods, analyse their results and leverage Julia's parallelism capabilities to perform Global Sensitivity analysis at scale.
using GlobalSensitivity, QuasiMonteCarlo, OrdinaryDiffEq, Statistics, WGLMakie
function f(du,u,p,t)
du[1] = p[1]*u[1] - p[2]*u[1]*u[2] #prey
du[2] = -p[3]*u[2] + p[4]*u[1]*u[2] #predator
end
u0 = [1.0;1.0]
tspan = (0.0,10.0)
p = [1.5,1.0,3.0,1.0]
prob = ODEProblem(f,u0,tspan,p)
t = collect(range(0, stop=10, length=200))
f1 = function (p)
prob1 = remake(prob;p=p)
sol = solve(prob1,Tsit5();saveat=t)
[mean(sol[1,:]), maximum(sol[2,:])]
end
bounds = [[1,5],[1,5],[1,5],[1,5]]
reg_sens = gsa(f1, RegressionGSA(true), bounds)
fig = Figure(resolution = (600, 400))
ax, hm = WGLMakie.heatmap(fig[1,1], reg_sens.partial_correlation, figure = (resolution = (600, 400),), axis = (xticksvisible = false,yticksvisible = false, yticklabelsvisible = false, xticklabelsvisible = false, title = "Partial correlation"))
Colorbar(fig[1, 2], hm)
ax, hm = WGLMakie.heatmap(fig[2,1], reg_sens.standard_regression, figure = (resolution = (600, 400),), axis = (xticksvisible = false,yticksvisible = false, yticklabelsvisible = false, xticklabelsvisible = false, title = "Standard regression"))
Colorbar(fig[2, 2], hm)
fig
using StableRNGs
_rng = StableRNG(1234)
morris_sens = gsa(f1, Morris(), bounds, rng = _rng)
fig = Figure(resolution = (600, 400))
scatter(fig[1,1], [1,2,3,4], morris_sens.means_star[1,:], color = :green, axis = (xticksvisible = false, xticklabelsvisible = false, title = "Prey",))
scatter(fig[1,2], [1,2,3,4], morris_sens.means_star[2,:], color = :red, axis = (xticksvisible = false, xticklabelsvisible = false, title = "Predator",))
fig
sobol_sens = gsa(f1, Sobol(), bounds, N=5000)
efast_sens = gsa(f1, eFAST(), bounds)
fig = Figure(resolution = (600, 400))
barplot(fig[1,1], [1,2,3,4], sobol_sens.S1[1, :], color = :green, axis = (xticksvisible = false, xticklabelsvisible = false, title = "Prey (Sobol)", ylabel = "First order"))
barplot(fig[2,1], [1,2,3,4], sobol_sens.ST[1, :], color = :green, axis = (xticksvisible = false, xticklabelsvisible = false, ylabel = "Total order"))
barplot(fig[1,2], [1,2,3,4], efast_sens.S1[1, :], color = :red, axis = (xticksvisible = false, xticklabelsvisible = false, title = "Prey (eFAST)"))
barplot(fig[2,2], [1,2,3,4], efast_sens.ST[1, :], color = :red, axis = (xticksvisible = false, xticklabelsvisible = false))
fig
fig = Figure(resolution = (600, 400))
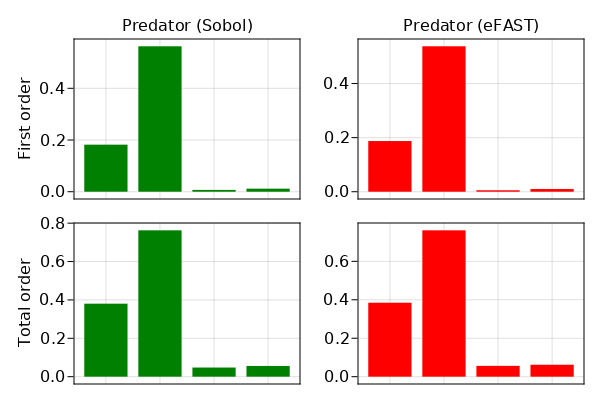
barplot(fig[1,1], [1,2,3,4], sobol_sens.S1[2, :], color = :green, axis = (xticksvisible = false, xticklabelsvisible = false, title = "Predator (Sobol)", ylabel = "First order"))
barplot(fig[2,1], [1,2,3,4], sobol_sens.ST[2, :], color = :green, axis = (xticksvisible = false, xticklabelsvisible = false, ylabel = "Total order"))
barplot(fig[1,2], [1,2,3,4], efast_sens.S1[2, :], color = :red, axis = (xticksvisible = false, xticklabelsvisible = false, title = "Predator (eFAST)"))
barplot(fig[2,2], [1,2,3,4], efast_sens.ST[2, :], color = :red, axis = (xticksvisible = false, xticklabelsvisible = false))
fig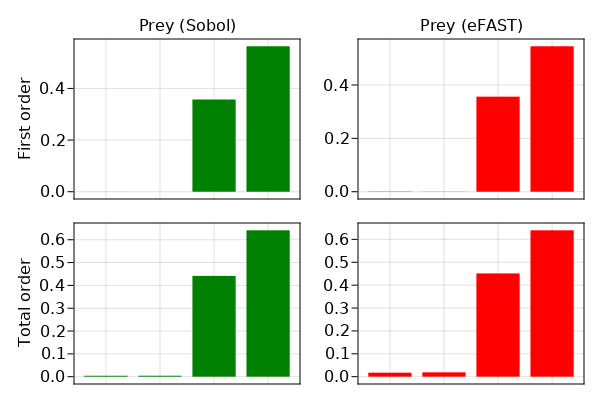
using QuasiMonteCarlo
N = 5000
lb = [1.0, 1.0, 1.0, 1.0]
ub = [5.0, 5.0, 5.0, 5.0]
sampler = SobolSample()
A,B = QuasiMonteCarlo.generate_design_matrices(N,lb,ub,sampler)
sobol_sens_desmat = gsa(f1,Sobol(),A,B)
f_batch = function (p)
prob_func(prob,i,repeat) = remake(prob;p=p[:,i])
ensemble_prob = EnsembleProblem(prob,prob_func=prob_func)
sol = solve(ensemble_prob, Tsit5(), EnsembleThreads(); saveat=t, trajectories=size(p,2))
# Now sol[i] is the solution for the ith set of parameters
out = zeros(2,size(p,2))
for i in 1:size(p,2)
out[1,i] = mean(sol[i][1,:])
out[2,i] = maximum(sol[i][2,:])
end
out
end
sobol_sens_batch = gsa(f_batch,Sobol(),A,B,batch=true)
@time gsa(f1,Sobol(),A,B)
@time gsa(f_batch,Sobol(),A,B,batch=true)